Tommy Rodriguez
Department of Research & Development, Pangaea Biosciences, Miami, FL, USA
Email: trodriguez[@]pangaeabio.com
Rodriguez, T. (2017). Phylogenetic Analysis of Maternal Lineages in Modern-Day Breeds of British Canis lupus familiaris International Journal of Research Studies in Biosciences 2017,5(9) : 41-47. DOI: http://dx.doi.org/10.20431/2349-0365.0509008
Abstract
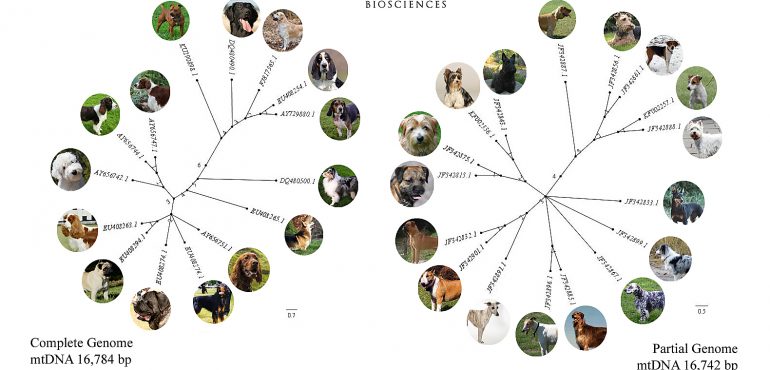
Domesticated dogs are byproducts of controlled breeding practices that make it difficult to establish a historically accurate phylogeny of divergent events among any localized group. Despite the difficulties of arriving at an accurate inference, this study found distinct phylogenetic clusters within modernday British dogs that largely corresponded to phenotype or function. Here, I examine maternal lineages among post-Victorian dog breeds by way of mitochondrial DNA (mtDNA) sequences. Two distinct sets of raw sequences were used in this investigation. My results will show that modern-day hounds, herding breeds, and spaniels fall nearest the midpoint of unrooted trees and contain the highest nucleotide substitution rates in the entire network, making these particular varieties strong candidates for the closest living relatives to the oldest lineages of European ancestry in Britain.
Keywords: domestic dog, phylogenetics, multiple sequence alignment, pairwise sequence alignment