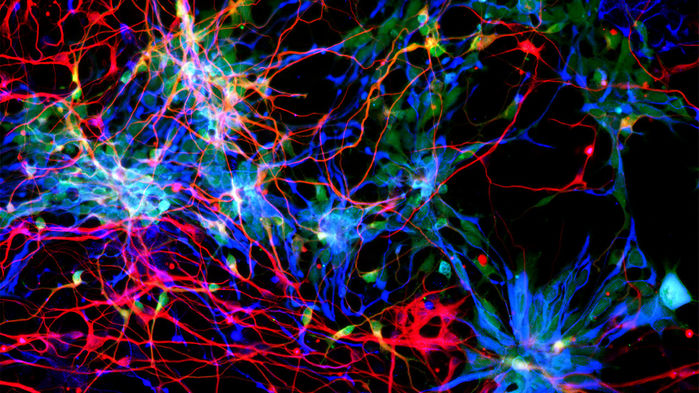
Three nearly identical genes could help explain how 0.5 liters of gray matter in early human ancestors became the 1.4-liter organ that has made our species so successful and distinctive. The newly identified genes could also help explain how brain development sometimes goes wrong, leading to neurological disorders.
The genes, descendants of an ancient developmental gene that multiplied and changed in the course of evolution, add to a growing list of DNA implicated in human brain expansion. But they stand out because so much has been learned about how they work their magic, says James Noonan, an evolutionary genomicist at Yale University. Researchers have shown that this trio boosts the number of potential nerve cells in brain tissue, and one team even pinned down the protein interactions likely responsible. “These are new proteins that are potentially modifying a very important pathway in brain development in a very powerful way,” Noonan adds.
Until now, the four genes were thought to be one, NOTCH2NL, itself a spinoff of the NOTCH gene family, which controls the timing of development in everything from fruit flies to whales. But two studies in the 31 May issue of Cell trace a series of genetic accidents in recent evolutionary history that have yielded four very closely related NOTCH2NL genes in humans (see graphic, below).
David Haussler, a bioinformatician at the University of California, Santa Cruz, and his colleagues got on the trail of the genes after they discovered that the NOTCH pathway works differently in human and macaque brain organoids—test tube models of the developing brain. NOTCH2NLwas missing in the macaque organoid and, later analyses showed, in other nonhuman apes as well. That suggested NOTCH2NL might have played a unique role in human evolution.
By comparing NOTCH2NL-related DNA in the genomes of humans and other primates, Haussler’s team reconstructed the genes’ evolutionary history. They concluded that during DNA replication perhaps 14 million years ago, part of an ancestral NOTCH2 gene was copied by mistake. The new “gene” was incomplete and nonfunctional, but about 11 million years later—shortly before human ancestors’ brains began to expand—an additional piece of NOTCH2 got inserted into this copy, making the gene functional. “This event marks the birth of the NOTCH2NL genes we now have in our brains,” says Frank Jacobs, a co–senior author on the paper and an evolutionary genomicist at the University of Amsterdam.
Subsequently, that active NOTCH2NL gene was duplicated twice more, yielding three active NOTCH2NL genes in a row at one end of human chromosome 1 and one inactive copy on the other end. Gene copies can be potent evolutionary forces because one copy continues its necessary job, leaving the others free to do something new.
Pierre Vanderhaeghen, a developmental neurobiologist at the Free University of Brussels, uncovered the same set of genes when he found a way to screen human fetal brain tissue for duplicated genes. To find out what they do, his team ramped up NOTCH2NL activity in cultured brain tissue. The tissue made more stem cells, they report in the second Cell paper.
The finding complements one reported earlier this spring by Wieland Huttner, a neurobiologist at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany. He and his team had decided to focus on NOTCH2NL (which they thought was a single gene) after finding it was highly active in fetal brain cells. When they put a human NOTCH2NL gene into incipient brain tissue from mice embryos, more stem cells developed. That suggests the human gene delays the specialization of those cells so they have a chance to produce many more copies of themselves, the researchers reported in eLife on 21 March.
Now, in their Cell paper, Vanderhaeghen and his colleagues describe molecular details of how NOTCH2NL works to boost neuron formation. They found that a NOTCH2NL protein blocks a key step in a signaling pathway that causes stem cells to differentiate and stop dividing. As a result, the cells persist and keep producing progeny, ultimately yielding a larger crop of neurons. “That’s really compelling biological data,” Noonan says. “In other studies of genes involved in human evolution, it’s been very difficult to draw a line from the genetic difference to the phenotype to a biochemical mechanism that’s responsible.”
The location of the three active NOTCH2NL genes is also telling, Haussler says. They are smack in the middle of DNA implicated in autism, schizophrenia, and a developmental delay syndrome. Such duplicated DNA is prone to getting copied extra times or losing DNA during replication, and instability is a hallmark of these disorders. To Greg Wray, an evolutionary developmental biologist at Duke University in Durham, North Carolina, this clue to brain diseases is the most compelling new result. “These genes likely play an important role in cortical development, and misregulation leads to disease,” he says.
Wray is less convinced that the genes had a unique role in human evolution because the chromosomal region in which they reside is complex and difficult to sequence, and because the evidence for an evolutionary difference in gene function between humans and other species is indirect.
But Haussler thinks these genes will prove key players in human brain expansion. “One change didn’t do it alone, but some will be found to be more fundamental than others,” he points out. “NOTCH2NL has a shot at this.”
Source: AAAS, Full Article